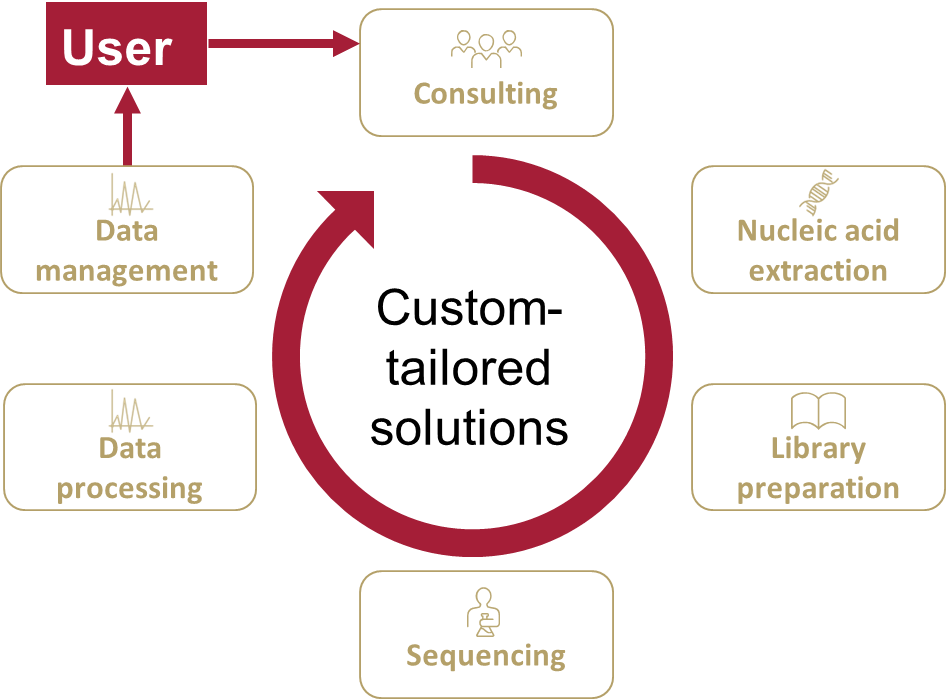
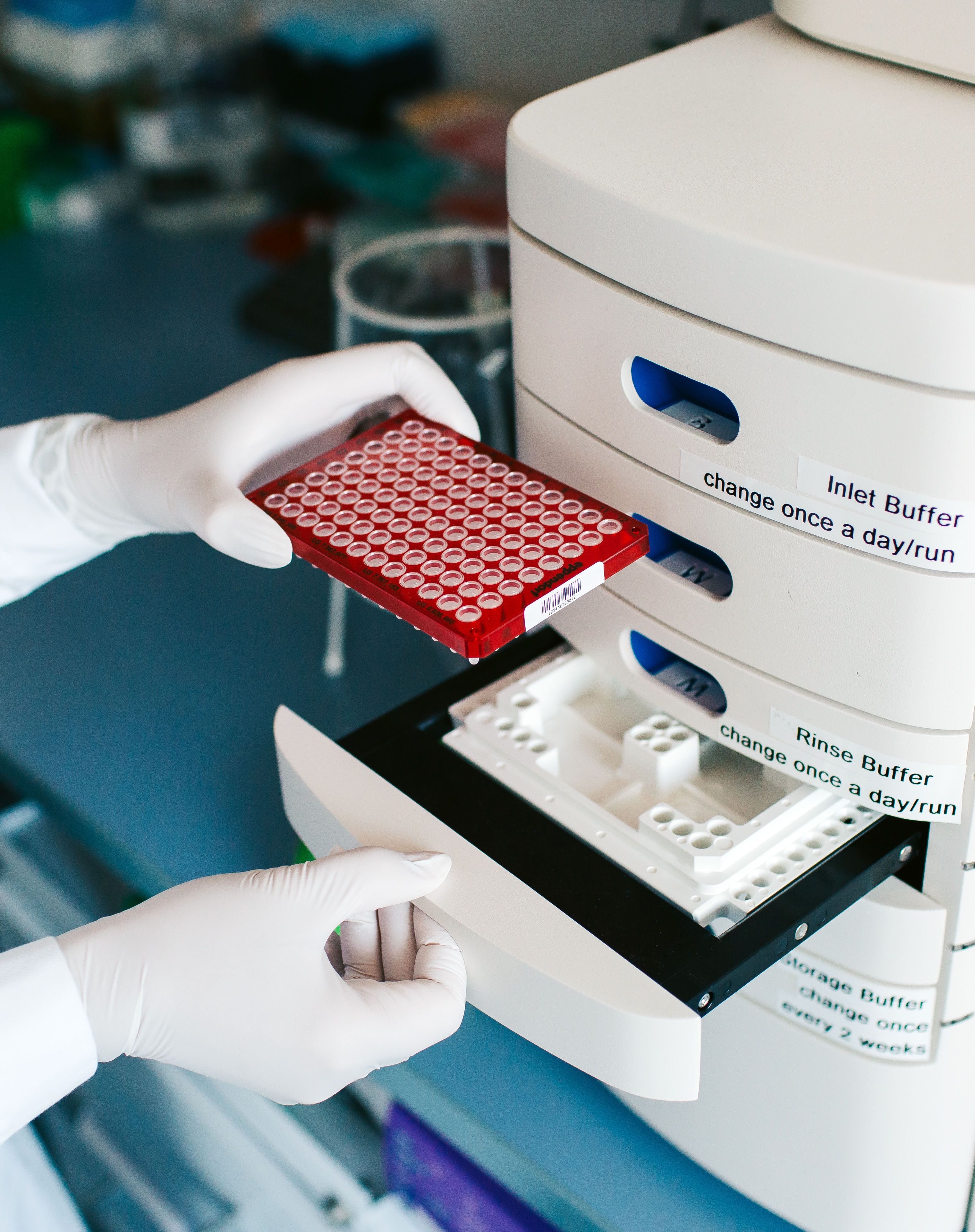
The Core Facility Genomics provides a wide range of next generation sequencing (NGS) methodologies for short- and long-read sequencing, along with tailored solutions for specific research needs. Our services prioritize expertise, reliability, and reproducibility, aiming to streamline functional genomics studies. Combining expertise from two specialized labs - human genetics (eukaryotes) and microbial genomics (prokaryotes) - we offer complementary resources, including biosafety and automation capabilities. Beyond sequencing, we support genomic research, microbial studies, model systems, and data science, focusing on custom solutions and advanced support for projects of all sizes.
About us
News
Services
 Consulting
Consulting
- For newcomers:
Which tool / which question - For experts:
Latest methods & benchmarking - For optimization:
Experimental design
 Nucleic acid extraction
Nucleic acid extraction
- DNA/RNA
- Diverse sample sources
 Library preparation
Library preparation
- Whole genome sequencing
Short/long read - Single cell sequencing:
Gene expression, immune profiling, chromatin, antibody tag-based proteomics - Targeted sequencing:
Whole exome, panels, amplicon - RNA sequencing:
TotalRNA, mRNA, smallRNA, low input, RNA isoform, directRNA - Metagenomics:
Shotgun, amplicons 16S, ITS - Circulating DNA
- Epigenomics:
Methylation - Metatranscriptomics
 Sequencing
Sequencing
- Illumina and ONT sequencing options
- QC and flow cells for ready-to-sequence libraries/pools
 Data processing and management
Data processing and management
- Sequencing QC
- Demultiplexing
- Based on research collaboration:
- Mapping in reference genomes
- Variant calling
- Gene expression - Further analyses and data storage options are available via QBiC
Teams

Human Genetics Group
Address:
Calwerstraße 7
721076 Tübingen
Phone number: +49 7071 29-72298
E-mail address: ncct@med.uni-tuebingen.de
Institution: How to find us

Dr. Nicolas Casadei
Head of Core Facility Genomics

Dr. Elena Buena Atienza
Scientist Long Reads

Dr. Sabine Fraschka
Scientist Single Cell

Michaela Pogoda, M.Sc
Scientist Short Reads, Automation

Jakob Admard, M.Sc
Scientist Bioinformatics

Mehmet Direnc Mungan
Scientist Bioinformatics

Sven Poths
Laboratory Manager

Jessica Klaus
Technician Short Reads

Noella Ansorge
Technician Long Reads

Antje Schulze-Selting
Technician Single Cell

Microbial Genomics Group
Address:
Elfriede-Aulhorn-Straße 6
72076 Tübingen
Phone number: +49 7071 29-61951
E-mail address: ncct@med.uni-tuebingen.de
Institution: How to find us

Dr. Janina Geißert-Janek
Deputy Head of Core Facility Genomics
Scientist

Dr. Janina Vogt
Team leader microbiology lab, Scientist

Dr. Oleksandra Karpiuk
Scientist

Dr. Miriam Lucke
Scientist

Jennifer Müller, M.Sc
Scientist Bioinformatics

Dr. Andrea Borbon-Garcia
Scientist Bioinformatics

Manuela Löffler
Technician

Chukwunonso Nnabude
Technician
Teams 2

Sven Dobler
Finance Management
Equipment
Sequencers
Small scale, medium scale and high-throughput sequencers are available from Illumina and Oxford Nanopore Technologies, supporting short-read and long-read technology.
Images are courtesy of Illumina, Inc. and Oxford Nanopore Technologies.

Illumina MiSeq
- Flexible low-output sequencer
- Read length: 50-600 bp
- Output: 0.54 - 15 Gb

Illumina NextSeq 500/550
- Mid-output sequencer
- Read length: 75 -300 bp
- Output: 16.25 - 120 Gb

Illumina NovaSeq 6000
- High-output sequencer
- Read length: 35 - 500 bp
- Output: 80 - 6,000 Gb

Illumina NovaSeq X Plus
- High-output sequencer
- Read length: 50 - 300 bp
- Output: 165 - 16,000 Gb

Oxford Nanopore MinION
- Low-throughput long-read sequencer
- Output: application dependent 300 Mb - 10 Gb / flow cell

Oxford Nanopore PromethION
- High-throughput long-read sequencer
- Output: application dependent 30 - 120 Gb / flow cell
Liquid Handling
Liquid handling systems help to increase reproducibility and scale up production. Several platforms for nucleic acid extraction and library preparation are implemented.
Tecan Fluent 780
High-throughput liquid handler
Implemented on site for bacterial genomic applications
Beckman i5
High-throughput liquid handler
Implemented on site for pre-PCR eukaryotic applications
Beckman i7
High-throughput liquid handler
Implemented on site for pre-PCR and post-PCR eukaryotic applications
QIAsymphony
High-throughput nucleic acid extraction
DNA & RNA isolation
QIAcube HT
High-throughput nucleic acid extraction
DNA & RNA isolation
Specific Equipment
Qubit
Fluorometer for DNA/RNA quantification
Bioanalyzer
Microfluidic device for DNA/RNA quantification and quality control
FemtoPulse
Automated pulse-field capillary electrophoresis system to assess quality and quantity of
high molecular weight DNA, as well as very low input DNA
TapeStation
Automated capillary electrophoresis system to assess quality and quantity of DNA or RNA samples
10x ChromiumX
High-throughput automated single-cell partitioning
10x CytAssist (for Visium HD)
Automated diffusion system to improve the resolution of spatial transcriptomics
FAQ
How do I get started?
Please contact us if we can support you in the planning of your sequencing project.
Before reaching out, please collect the following data:
- What is the aim of your project?
- How many samples do you have?
- How many replicates are planned?
- What is your sample type? (e.g. library, DNA/RNA, blood, stool, etc.)
- Are your samples genetically modified organisms (GMOs), do they present a biosafety risk?
- Which species are you working on? (e.g. human, mouse, etc.)
- Is bioinformatics analysis needed? If yes, to what degree?
You can provide your sample details in our Metadata Sheet.
Please enter your contact and sample details and submit in digital form and in printed version together with your samples.
What are the sample requirements?
Sample guidelines, shipment address and other useful information can be found on this page under resources.
Could I bring the samples personally?
Yes, it is possible. Please contact us beforehand to coordinate date and time. Even if the travel time is short, please ensure that the samples are stored properly during transportation (e.g., they are frozen or kept on ice).
Do I have to use dry ice for RNA sample shipment?
The quality of RNA and subsequent RNA-seq is strongly dependent on its storage and transportation conditions, so we strongly encourage using dry ice for RNA shipment. If you are shipping samples for RNA isolation, they also have to be shipped on dry ice. Important note: samples in RNA stabilizing solutions (e.g., RNAlater) can be shipped chilled or frozen at -20°C. Please notify us beforehand if your samples contain RNA stabilizing solution.
What is the sample turnaround time?
Without prior information about your project it is very difficult to estimate the time required for it. The turnaround time depends on sample number, exact sample prep workflow, equipment and personnel availability. Please contact us for more details. Please note, during winter and summer holidays, the turnaround time is slower due to personnel availability.
Which data do I get?
The standard output from a sequencing project is demultiplexed FASTQ files. If further bioinformatics support is needed, please contact us.
Is there a price list available?
Please contact us to receive a quote for your project.