Division of Translational Oncology
Main focus of our laboratory is to understand the mechanisms of hematopoietic differentiation and leukemogenic transformation. We apply multidisciplinary approaches covering different research fields.
Main focus of our laboratory is to understand the mechanisms of hematopoietic differentiation and leukemogenic transformation. We apply multidisciplinary approaches covering different research fields.

Prof. Dr. med., Ph.D. Julia Skokowa
W3 Professur für Translationale Onkologie
Publications: Publications
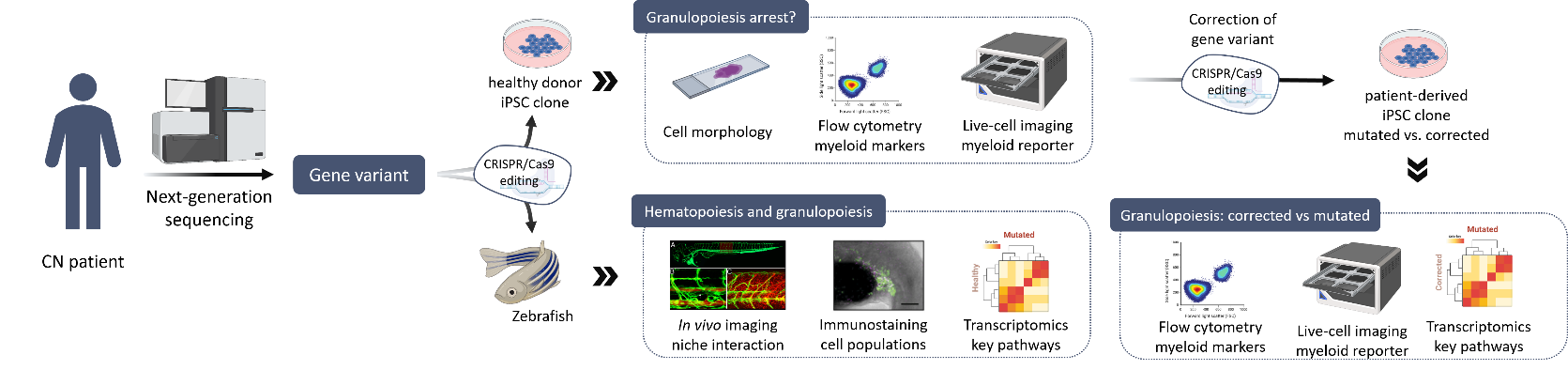
Severe congenital neutropenia (CN) is a rare heterogeneous bone marrow syndrome characterized by maturation arrest of granulopoiesis at the level of promyelocytes/myelocytes, resulting in an absence of mature granulocytes in peripheral blood (neutropenia) and severe recurrent bacterial infections. So far, more than 20 different genes have been identified to be associated with CN, including ELANE, HAX1, and JAGN1; however, their pathophysiological mechanisms are not fully understood. Moreover, almost 25% of CN patients remain without a genetic diagnosis.
In collaboration with the Severe Chronic Neutropenia International Registry (SCNIR), our group aims to identify and characterize novel gene mutations in patients with genetically unclassified CN. First, a comprehensive analysis of the genome using a next-generation sequencing pipeline for genetic diagnosis is completed. If a candidate gene variant is identified, the pathogenesis of the mutation is investigated using different experimental models. These models include studies in i) human hematopoietic stem cells, ii) induced pluripotent stem cell (iPSC) derived from healthy individuals and CN patients, and iii) zebrafish. Overall, these studies allow us to determine whether the new variant is responsible of the maturation arrest of granulopoiesis and to further uncover downstream mechanisms involved in the pathogenesis. Then, we further work to define the pathomechanisms of CN-associated genes in disease development in order to better understand disease development and define therapeutic targets.
CN is a pre-leukemia syndrome with a cumulative incidence of myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) of more than 20% after 20 years. The progression of CN to MSD and/or AML is a multistep process. HSPCs harboring inherited mutations acquire early driver mutations during neutropenic phase (e.g., CSF3R mutations), followed by additional mutations before onset of full-blown MDS/AML.
In order to improve early diagnosis and treatment of CN patients that may develop leukemia, our group aims to identify additional gene mutations in leukemia-associated genes in CN-MDS/AML patients using a NGS-based sequencing panel of leukemia-associated genes and comparing CN/AML patient samples to CN samples in order to identify additional candidate mutations. To further characterize these leukemia-associated mutations in CN-MDS/AML patients, we use different experimental models to assess the leukemogenic potential of new mutations in malignant transformation and uncover intracellular signaling pathways involved in disease development.
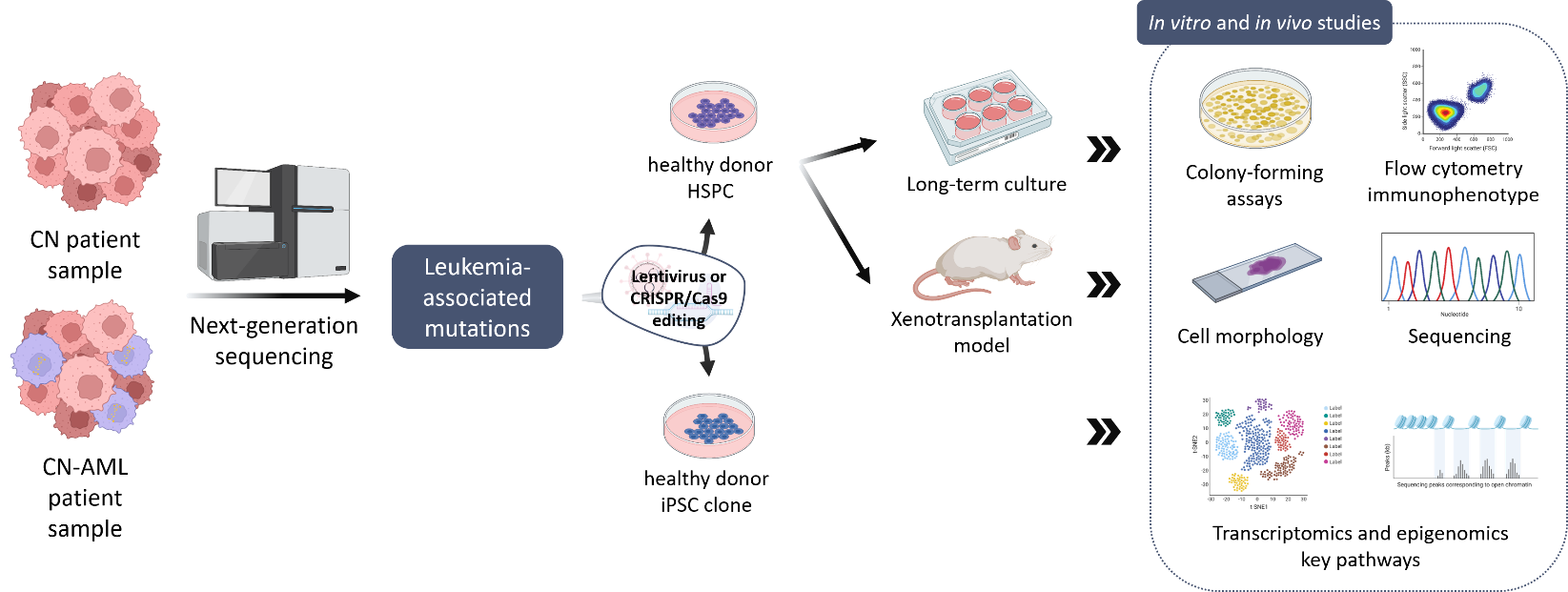
Our group established a disease model of stepwise leukemogenesis in CN/AML by CRISPR-Cas9 gene editing of CN/AML patient-derived iPSCs and subsequent hematopoietic differentiation. This model we also used for AML drug discovery. We identified BAALC upregulation and the following phosphorylation of MK2a as a key leukemogenic event. BAALC deletion or treatment with CMPD1, a selective inhibitor of MK2a phosphorylation, blocked proliferation and induced differentiation of primary CN/AML blasts and CN/AML iPSC-derived hematopoietic stem and progenitor cells (HSPCs) without affecting healthy donor or CN iPSC-derived HSPCs. Our disease model is suitable for future investigation of stepwise leukemogenesis and AML drug screening. This study suggests that targeting BAALC and/or MK2a phosphorylation may prevent leukemogenic transformation or eliminate AML blasts in CN/AML and RUNX1-mutant BAALChigh de novo AML. (Dannenmann B, Klimiankou M et al. Cell Stem Cell. 2021)

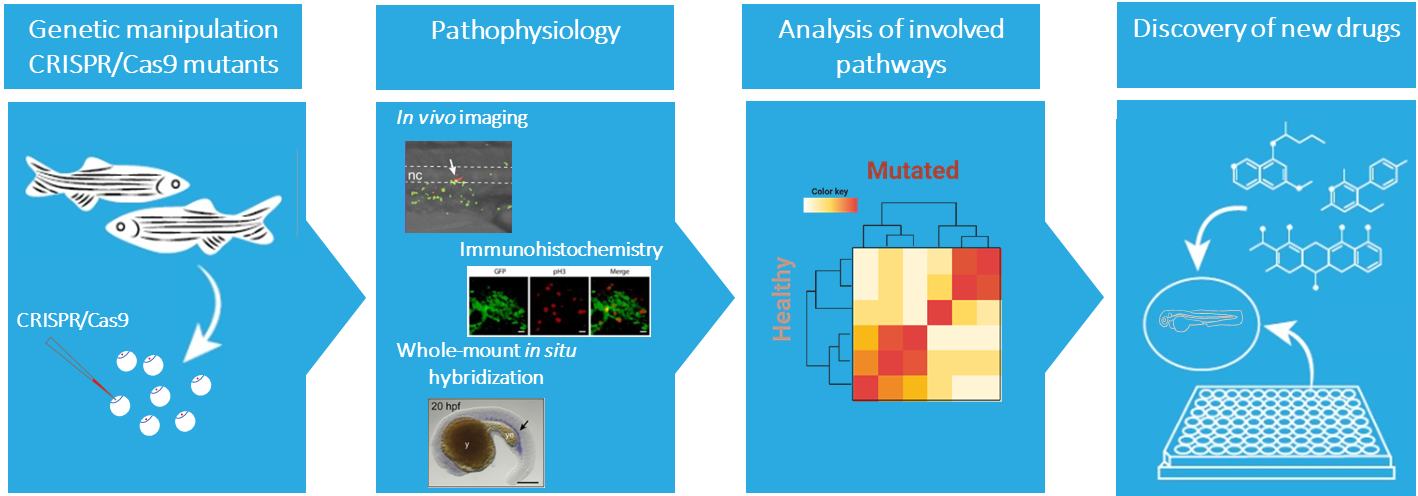
The information acquired from model organisms is essential for understanding the development of a disease. The zebrafish (Danio rerio) is an attractive complementary model system. The molecular and cellular mechanism underlying hematopoiesis are largely conserved between zebrafish and humans, making the zebrafish a very suitable vertebrate model to study normal and malignant blood development. Also, the optical transparency and ex vivo development of the embryos and larvae allows monitoring throughout embryogenesis, providing unique accessibility to embryonic lethal mutations. Additionally, the availability of fluorescent transgenic reporter lines allows an easy observation of different cell behavior and analysis of niche interactions.
Our current knowledge on genes associated with severe congenital neutropenia is predominantly derived from clinical observations and in vitro studies. In our group we use zebrafish as a model to fill the gap and understand the molecular mechanisms underlying congenital neutropenia and leukemia development. In addition, the zebrafish will serve as an in vivo platform to identify new avenues for developing tailored therapeutic strategies for patients for various types of blood malignancies, including specific subtypes of ALL, AML, MDS, and MM as well as inherited pre-leukemic bone marrow failure syndromes.
Our protein design team develops and deploys cutting-edge design methods to generate novel proteins for biomedical applications. For evaluation of the designed molecules, we characterize their biophysical properties, determine their structures, and evaluate their activity in vitro, ex vivo and in vivo assays. Our research mainly focuses on the following major areas:
Despite recent advances in computational protein design, simultaneous enhancements of design throughput and accuracy are continually needed to achieve better success rates at the bench, and tackle more complex design problems. We are continually developing the Damietta design engine; a design framework that leverages the accuracy of established molecular mechanics force fields, while maximizing the throughput of energy calculations through tensorized implementations. While still a bleeding edge technology, we have already successfully used the Damietta engine for different design projects in the lab. Other development domains cover ultra-fast docking and shape similarity algorithms, that enable us to design de novo and rescaffolded binders.
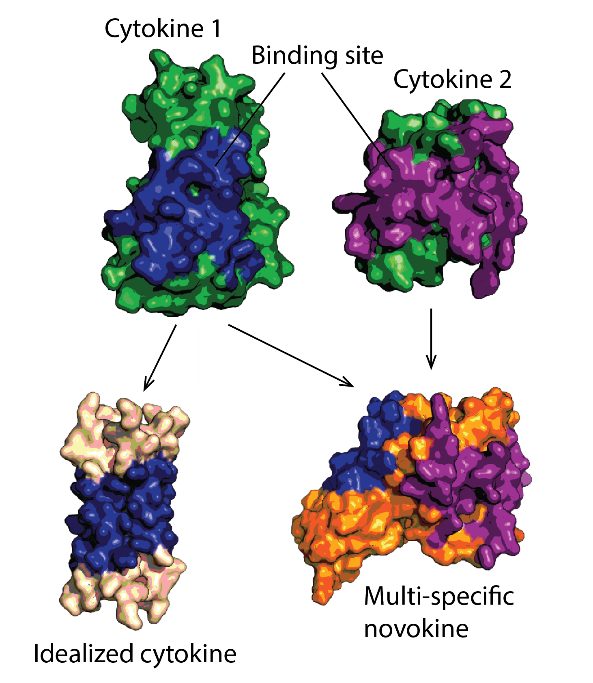
Protein therapeutics must meet a range of requirements to their activity, stability, solubility, aggregation propensity, and production costs. Computational design can offer control over these parameters and greatly facilitate protein drug development. We use a set of in-house developed computational methods for tertiary epitope matching, protein core re-design, interface design, loop design, and accelerated dynamics scoring in order to make idealized therapeutic miniproteins. We design and preclinically-develop idealised growth factors and cytokines with superior properties compared to their natural counterparts. We also use these designs to study the structure-function relationship of receptor modulation. Furthermore, we design novel-function cytokines (i.e. novokines) with the goal of inducing novel signaling patterns in target cells, at a very high targeting specificity.
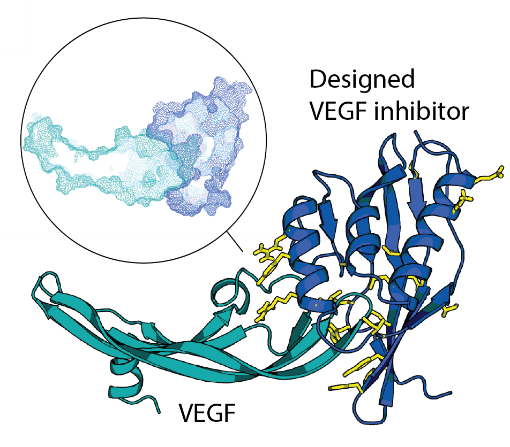
De novo design of site-specific binders in a concept-to-hit manner remains a major scientific and technological challenge. We established a novel framework to rapidly create epitope-directed binders using only information on the target structure. Our pipeline takes advantage of a high throughput docking software that evaluates steric complementarity of proteins in a lower dimensional space and enables selection of shape matching scaffolds for further interface design. Previously, we made inhibitors blocking an active site of VEGF, a key angiogenic molecule involved in the pathogenesis of diverse cancers, cardiovascular, and ophthalmic diseases. Moving forward, we work on improvements of the entire pipeline and on design of binders against other relevant targets.
Metalloproteins have a wide range of biomedical applications. For example, they can serve as electron microscopy contrast agents, as emergency antidotes for metal intoxication or as genetically-encodable protein tags for targeted radioimmunotherapy. We aim to design stable, easy-to-purify proteins that would bind copper, lead or other metals with high affinities and high metal binding capacity. For this, we use different approaches, from redesigning natural metalloproteins to mounting multiple metal binding sites onto novel geometrically-accommodating scaffolds.

Focus: Top National Hospital 2025

Stern: Germany's Outstanding Employers in Nursing 24/25

Quality partnership with the PKV

Family as a success factor

Pension provision for the public sector