AG Walz

Peptide-based Immunotherapy
The overall goal of the Department of Peptide-based Immunotherapy is the development of novel immunotherapy concepts for malignant and infectious disease. Based on this, our research activity and expertise can be divided into four key areas:
- The direct mass spectrometry-based analysis of the immunopeptidome and the characterization of the antigen-specific T cell repertoire of malignant disease. With the combination of these unique techniques we provide a leading platform for the identification and characterization of tumor-associated antigens and for the evaluation of the dynamic of the tumor immunopeptidome and peptide-specific T cell response under the influence of drugs and stress conditions.
- The characterization and optimization of antigen-specific T cell responses in infectious diseases.
- The non-GMP and GMP production of peptides and peptide-based vaccines
- The translation of novel immunotherapeutic strategies from bench to bedside with a focus on peptide-based immunotherapies with currently four ongoing clinical trials evaluating own-developed peptide vaccines
List of GRT projects
Monitoring of T cell immunity for Gene and RNA therapies
Within these projects we apply our expertise in the characterization of antigenspeciif T cells to monitor the immunogenicity of gene and RNA therapeutics. For mRNA-based immunotherapies/vaccines the induction of T cell response is central for the clinical success of the compound (Maringer et al., Science Immunol 2022). For gene or protein-based therapeutics on the other hand immunogenicity i.e., specific T cell responses targeting the respective compound should be avoided. High-throughput T cell epitope screening and multiparametric analysis of antigen-specific T cell response is based on our unique prediction and selection workflow, integrating the algorithms SYFPEITHI and NetMHCpan.
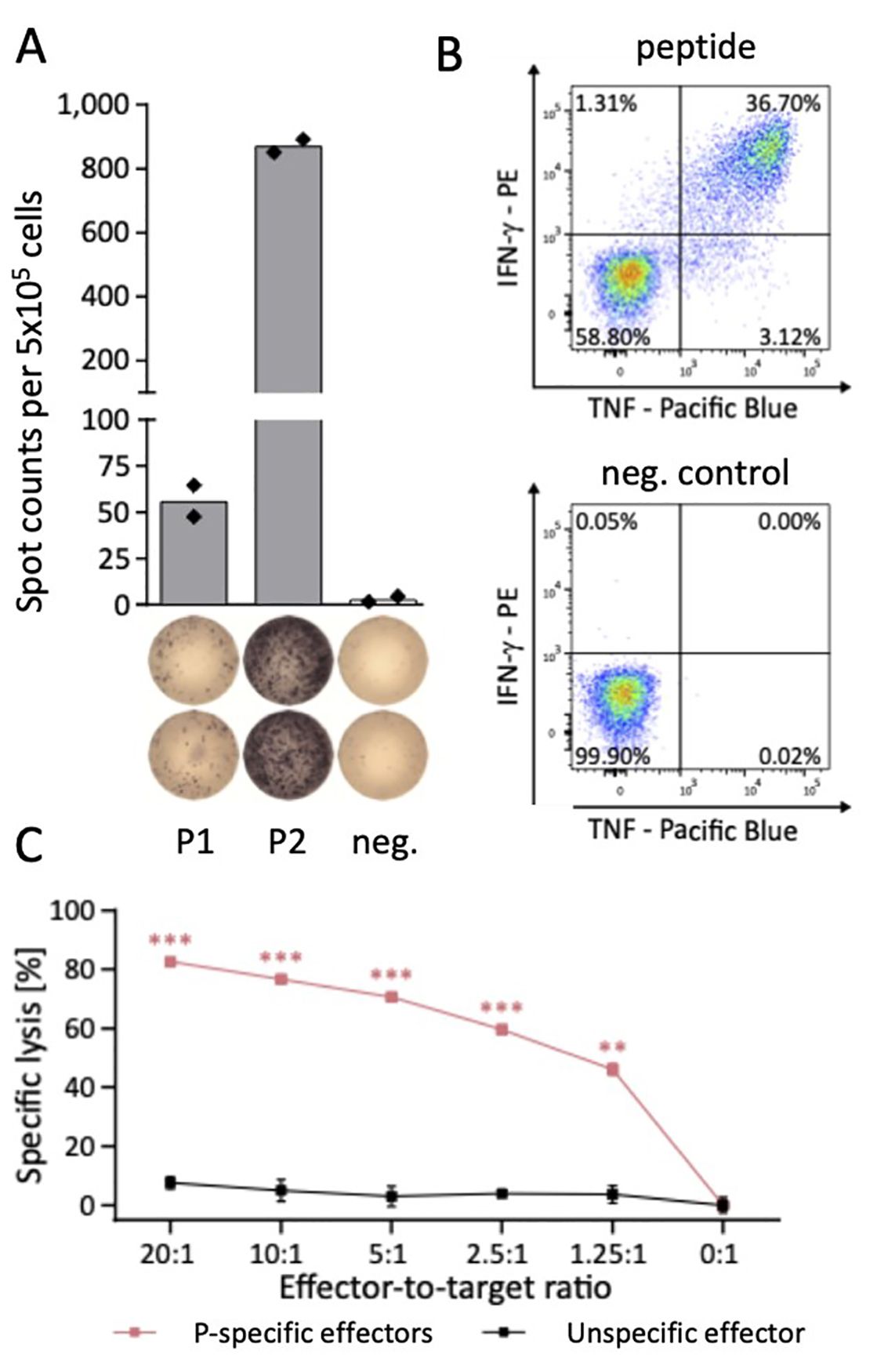
This workflow enables the identification of dominant and highly immunogenic CD8+ and CD4+ T cell epitopes. Immunogenicity screening T cell epitopes is performed by IFN-γ enzyme-linked immunospot (ELISPOT) representing a well-established, highly sensitive method for the high throughput screening of T cell epitopes (Fig. 1A).
To even detect low frequent antigen-specific T cells, we established a protocol to expand antigen-specific T cells in vitro prior to ELISPOT analysis. Beside high-throughput identification of T cell epitopes this approach further enables the standardized large-scale analysis and quantification of patient cohorts for functional T cell responses to multiple HLA-matching antigens38,44-46. For further multi-parametric characterization of these antigen-specific T cells we use a combinatorial approach of multi-parameter flow cytometry for T cell phenotype, T cell functionality and T cell exhaustion analysis (Fig. 1B) as well as two cytotoxicity assay systems (Vital assay, IncuCyte) evaluating specific lysis of peptide-loaded or peptide-presenting target cells by antigen-specific T cells (Fig. 1C).
GRT expertise
- Immunogenicity screening
- Immunopeptidome analysis
Main GRT methods applied in the lab
- IFN-g ELISPOT
- Multi color flow cytometry
- Cytotoxicity assays
- Mass spectrometry based immunopeptidomics
List/description of clinical trials
Please refer to
Drug peptide laboratory | Universitätsklinikum Tübingen (uni-tuebingen.de)
Ongoing:
- B-pVAC-SARS-CoV-2; Prof. Dr. Juliane Walz; University Hospital Tübingen
- iVAC-L-CLL01; Prof. Dr. Helmut Salih; University Hospital Tübingen
- iVAC-XS15-CLL01; Prof. Dr. Juliane Walz; University Hospital Tübingen
Completed:
- P-pVAC-SARS-CoV-2; Prof. Dr. Juliane Walz, University Hospital Tübingen (Heitmann JS et al, Nature)
Upcoming:
- FusionVac_22, Prof. Dr. Juliane Walz, University Hospital Tübingen
- AML_VAC_XS1501, Prof. Dr. Juliane Walz, University Hospital Tübingen
- PerVision, PD. Dr. Martin Ebinger; University Hospital Tübingen
Ongoing and requested funding
Research | Universitätsklinikum Tübingen (uni-tuebingen.de)
- Jose Carreras Foundation
- Wilhelm Sander Foundation
- Bundesministerium für Bildung und Forschung (BMBF)
- Ministerium für Wissenschaft, Forschung und Kunst Baden-Württemberg (MWK)
- Dieter Schwarz Stiftung
- Deutsche Krebshilfe
- Deutsches Konsortium für Translationale Krebsforschung (DKTK)
- Deutsche Forschungsgemeinschaft (DFG)
- Else Kröner Fresenius Stiftung
- iFIT Cluster of Excellence
Publications
- Maringer Y, Nelde A, Schroeder SM, Schuhmacher J, Hörber S, Peter A, Karbach J, Jäger E, Walz JS. Durable spike-specific T-cell responses after different COVID-19 vaccination regimens are not further enhanced by booster vaccination. Science Immunol. 2022 Nov 1:eadd3899. doi: 10.1126/sciimmunol.add3899; IF: 30.6
- Bauer J, Köhler N, Maringer Y, Bucher P, Bilich T, Zwick M, Dicks S, Nelde A, Dubbelaar M, Scheid J, Wacker M, Heitmann JS, Schroeder S, Rieth J, Denk M, Richter M, Klein R, Bonzheim I, Luibrand J, Holzer U, Ebinger M, Brecht IB, Bitzer M, Boerries M, Feucht J, Salih HR, Rammensee HG, Hailfinger S, Walz JS. The oncogenic fusion protein DNAJB1-PRKACA can be specifically targeted by peptide-based immunotherapy in fibrolamellar hepatocellular carcinoma. Nat Commun. 2022, 13, 6401. https://doi.org/10.1038/s41467-022-33746-3;IF 17.7
- Heitmann JS, Bilich T, Tandler C, Nelde A, Maringer Y, Marconato M, Reusch J, Jäger S, Denk M, Richter M, Anton L, Weber LM, Roerden M, Bauer J, Rieth J, Wacker M, Hörber S, Peter A, Meisner C, Fischer I, Löffler MW, Karbach J, Jäger E, Klein R, Rammensee HG, Salih HR, Walz JS. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 2022 Jan;601(7894):617-22. doi: 10.1038/s41586-021-04232-5; IF: 69.5
- Bilich T, Roerden M, Maringer Y, Nelde A, Heitmann JS, Dubbelaar MS, Peter A, Hörber S, Bauer J, Rieth J, Wacker M, Berner F, Flatz L, Held S, Brossart P, Märklin M, Wagner P, Erne E, Klein R, Rammensee HGR, Salih HR, Walz JS. Preexisting and Post-COVID-19 Immune Responses to SARS-CoV-2 in Patients with Cancer. Cancer Discov. 2021 Aug;11(8):1982-1995. doi: 10.1158/2159-8290.CD-21-0191; IF: 39.4
- Bilich T, Nelde A, Heitmann JS, Maringer Y, Roerden M, Bauer J, Rieth J, Wacker M, Peter A, Hörber S, Rachfalski D, Märklin M, Stevanović S, Rammensee HG, Salih HR, Walz JS. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci Transl Med. 2021 Apr 21;13(590):eabf7517. doi: 10.1126/scitranslmed.abf7517; IF: 18.0
- Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, Lübke M, Bauer J, Rieth J, Wacker M, Peter A, Hörber S, Traenkle B, Kaiser PD, Rothbauer U, Becker M, Junker D, Krause G, Strengert S, Schneiderhan-Marra N, Templin MF, Joos TO, Kowalewski DJ, Stos-Zweifel V, Fehr M, Rabsteyn A, Mirakaj V, Karbach J, Jäger E, Graf M, Gruber LC, Rachfalski D, Preuß B, Hagelstein I, Märklin M, Bakchoul T, Gouttefangeas C, Kohlbacher O, Klein R, Stevanović S, Rammensee HG, Walz JS. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T-cell recognition. Nature Immunol. 2021 Jan;22(1):74-85. doi: 10.1038/s41590-020-00808-x; IF: 31.2
- Bilich T*, Nelde A*, Bichmann L, Roerden M, Salih HR, Kowalewski DJ, Schuster H, Tsou CC, Marcu A, Neidert MC, Lübke M, Rieth J, Schemionek M, Brümmendorf TH, Vucinic V, Niederwieser D, Bauer J, Märklin M, Peper JK, Klein R, Kohlbacher O, Kanz L, Rammensee HG, Stevanović S, Walz JS. The HLA ligandome landscape of chronic myeloid leukemia delineates novel T-cell epitopes for immunotherapy. Blood. 2019 Feb 7;133(6):550-565. doi: 10.1182/blood-2018-07-866830; IF: 25.5
- Walz S*, Stickel JS*, Kowalewski DJ, Schuster H, Weisel K, Backert L, Kahn S, Nelde A, Stroh T, Handel M, Kohlbacher O, Kanz L, Salih HR, Rammensee HG, Stevanović S. The antigenic landscape of multiple myeloma: mass spectrometry (re)defines targets for T-cell based immunotherapy. Blood. 2015 Sep 3;126(10):1203-13. doi: 10.1182/blood-2015-04-640532; IF: 25.5
- Berlin C, Kowalewski DJ, Schuster H, Mirza N, Walz S, Handel M, Schmid-Horch B, Salih HR, Kanz L, Rammensee HG, Stevanović S, Stickel JS. Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. Leukemia. 2015 Mar;29(3):647-59. doi: 10.1038/leu.2016.1; IF: 12.9
- Kowalewski DJ, Schuster H, Backert L, Berlin C, Kahn S, Kanz L, Salih HR, Rammensee HG, Stevanović S, Stickel JS. HLA ligandome analysis identifies the underlying specificities of spontaneous anti-leukemia immune responses in chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci U S A. 2015 Jan 13;112(2):E166-75. doi: 10.1073/pnas.1416389112; IF: 11.2
Relevant links