The long-term research goal is to gain a detailed and comprehensive understanding of the role of the gut microbiota in health and disease.
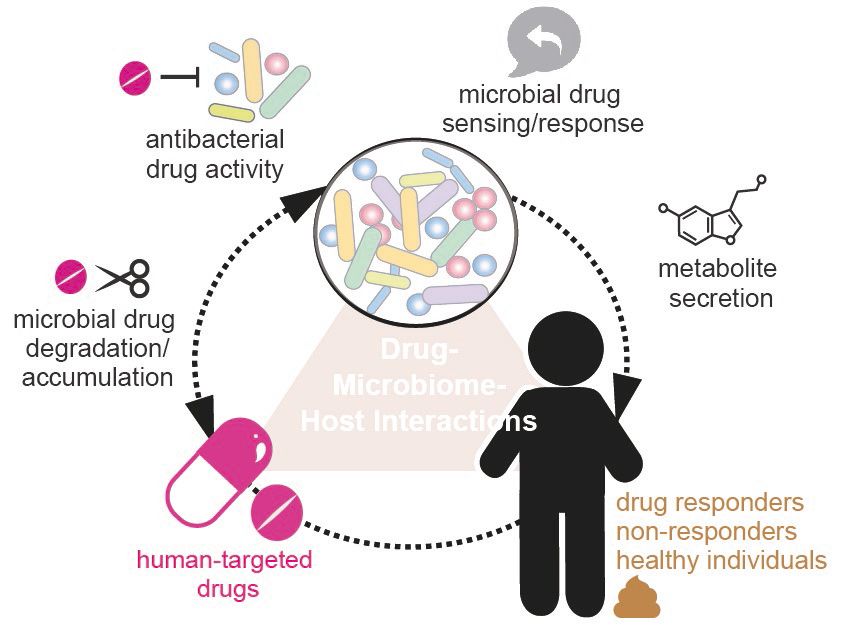
In recent years, we have developed a particular interest in the effects of medicinal drugs on the human gut microbiome. To understand the interactions between small molecules, the microbiome and the host, thelab uses microbiology-driven and cultivation-based bottom-up approaches. As a starting point, the team systematically maps the direct effects of drugs on gut bacteria in pure cultures, in microbial communities and in vivo. Based on the results of these systematic approaches, the team elucidates the underlying molecular mechanisms, investigates the development of resistance and explores the consequences for the host, such as drug efficacy, toxicity, pathogen colonisation or side effects. The current focus is on anticancer and psychotropic drugs, as these therapeutic drug classes show strong direct effects on the gut microbiome.
We are further using our insights into how small molecules modulate the gut microbiome to develop strategies for targeted pathogen decolonisation. For example, the lab aims to reverse ectopic colonisation of the gut with species from the oropharyngeal cavity - a scenario that has been shown to cause enteric environmental dysfunction (EED), is associated with chronic malnutrition in children and linked to colorectal cancer.
In addition, the lab is developing methods and protocols for isolating, culturing and investigating previously under-studied members of the human microbiome. This includes developing tools to illuminate the genomic dark matter of the human gut microbiome.