1. Artificial intelligence-based data processing
The application of AI methods in acquisition, reconstruction, post-processing or analysis are investigated. We develop reliable, robust, specific, and sensitive methods to serve this purpose. The inclusion of AI into medical data processing can help to improve performance by (but not limited to) increasing precision, boosting quality of service, easing processing, reducing computational times, and reducing energy consumption.
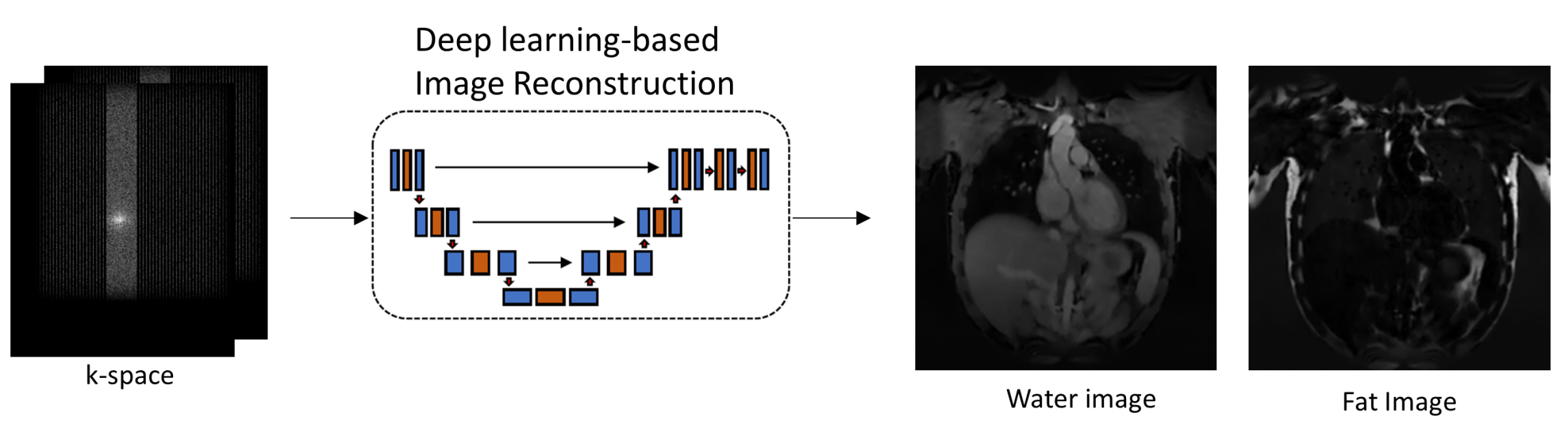
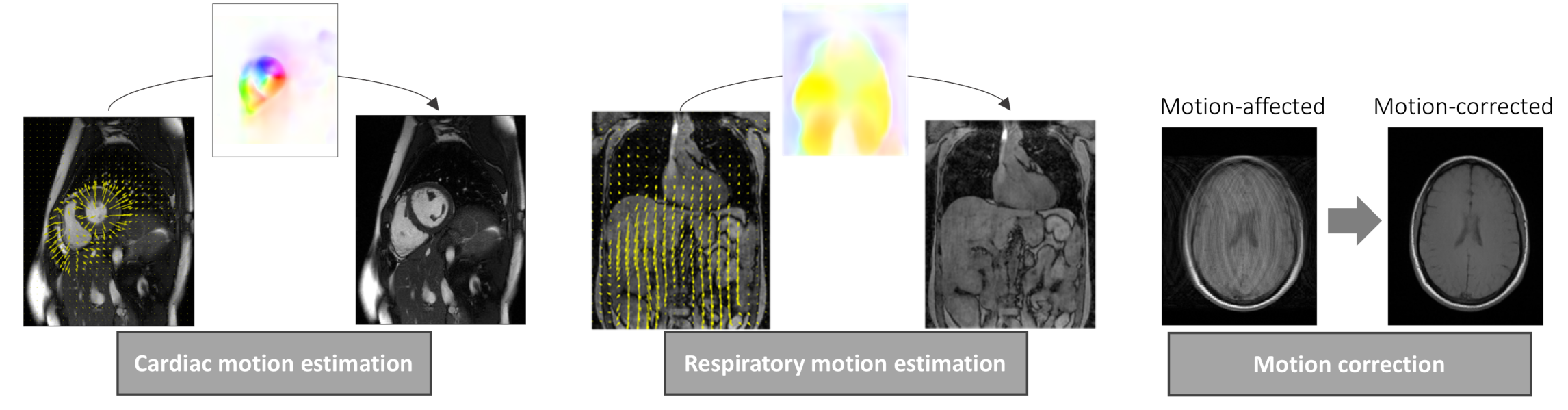
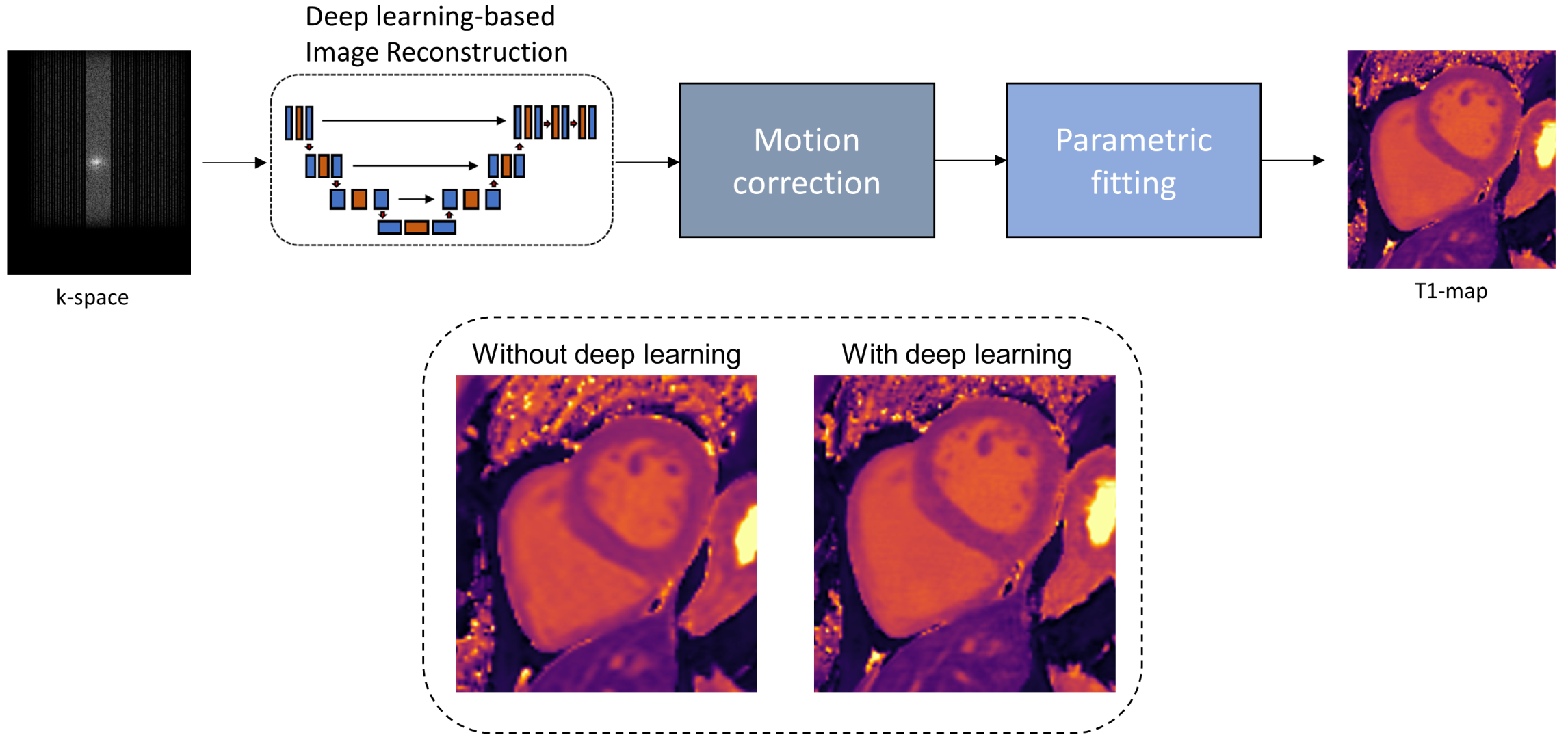
As the demand for medical imaging grows, the imaging-related energy consumption and sustainable operation is becoming a pressing concern for radiology departments and practices. The development of efficient acquisition strategies with multi-parametric and dynamic imaging is thus essential. In this regard, one-stop-shop imaging solutions and AI-based processing (reconstruction, handling of motion, image quality control) enable a more comprehensive non-invasive tissue and metabolism characterization.
The aim is to provide an improved workflow with automatic data handling to derive clinical biomarkers that can be used in diagnosis.
2. Patient-centered and epidemiological cohort analysis
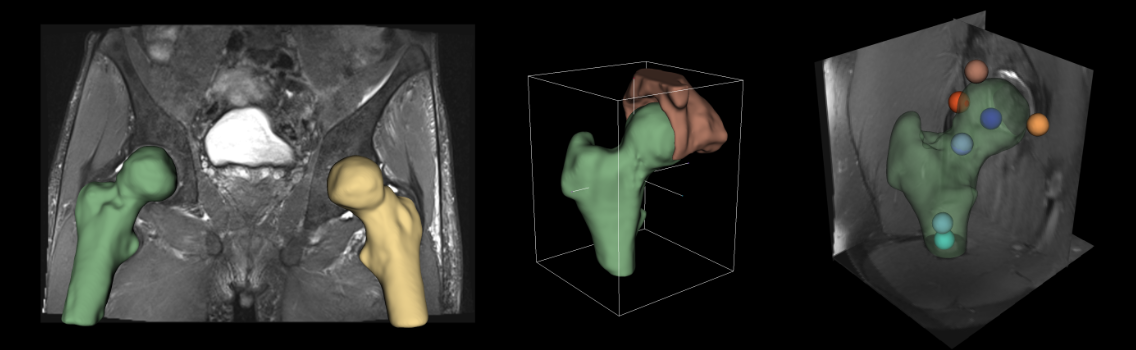
A patient-centered analysis combines information from imaging, genetics, laboratory data, meta data and expert knowledge to derive clinical biomarkers that can be used in diagnosis or therapy monitoring. In the context of large epidemiological studies, manual image analysis is often not feasible due to the overwhelming amount of data. The aim is to provide improved and automated workflows using AI-based processing while also investigating their role and impact. To this end, causal and explainable models are investigated for segmentation, volumetric measurements, textural composition, biological age estimation and treatment response prediction, in various organs, tissue and pathologies of interest. In order to guarantee a reliable image quality, data quality control checks and countermeasures are included. We investigate the causal relationships that lead to the AI-based findings and identify confounding factors with disentanglement of independent variables.
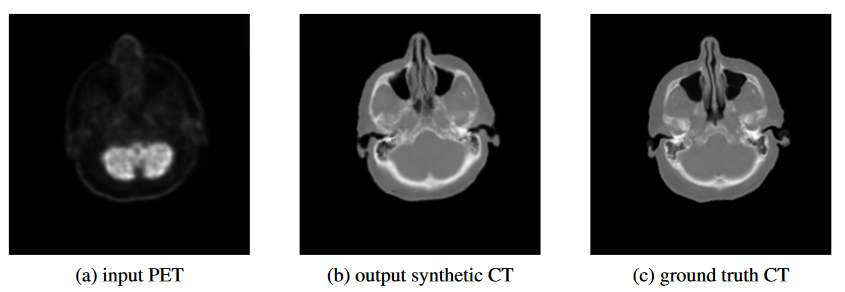
3. Translation of AI to clinical applications
We aim to implement state-of-the-art AI methods for clinical applications to support the clinicians in their daily work. These projects enable automated processing, simplified workflows and patient-phenotypic processing and analysis. Imaging and non-imaging data are coherently processed to guide and monitor patients with neurological, cardiovascular and oncological diseases. One example use-case is the automatic analysis of whole-body imaging data such as PET-CT. In this context we launched an ML challenge: autoPET.